Autonomous Science Part I: Everything’s an API Away

What would you build assuming AGI is a few years away?
A system capable of autonomously conducting novel science is at or near the top of our list. After investing in and studying the field for several years, we believe now is finally the time to work towards that future in which autonomous science will graduate from automating repetitive tasks and optimization problems to increasingly open-ended scientific exploration.
Building such a system is ultimately about the integration of technologies, each on their own exponential improvement curve. These include but aren’t necessarily limited to the ability to:
- Understand the current knowledge stored primarily in literature and datasets
- Reason upon that
- Come up with hypotheses to test or the next action to take
- Be able to carry out that experiment in the real world
- and/or conduct experimental exploration in silico whenever our digital tools are strong enough to yield reliable results
In Part I of our Automating Science posts, we detail the state of the art in these five technological categories, the trajectory they’re on, how their future developments may unfold, and how they interconnected.
Knowledge Graphs and Data Pools
The rapid growth of biomedical knowledge far outpaces our ability to efficiently extract insights and generate novel hypotheses. Publicly available literature includes 25M abstracts, 3M full texts, and 4k new papers a day. This is where we believe one could start in automating science.
Literature synthesis can be done sufficiently well by integrating LLMs with traditional lookup methods. At least before Deep Research, FutureHouse built the most clever technical implementation so far and then experimented with autonomously running their literature crawling system to passively screen the literature for contradictions to the claims made in the paper. Turns out roughly 2% of papers’ claims are debated by other papers.
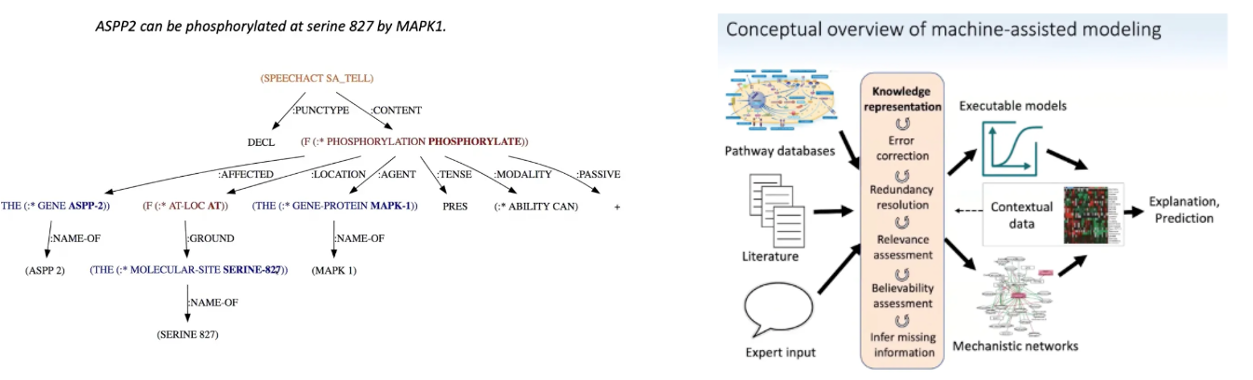
An even more ambitious project is a continually updating knowledge graph of individual fragments of mechanistic or causal knowledge – collected, curated and assembled with care to unify the literatures' claims.
An impressive attempt to do so at large scale came last year when researchers built an automated system that translates mined text into machine-readable causal mechanistic relationships in as much detail as possible. It then assembles and hierarchically organizes the fragments by handling errors in the AI’s reading comprehension, resolving redundancies and contradictions in the literature, understanding context, assessing a claim’s relevance and believability, inferring missing information, etc. It has analyzed ~1M papers so far.
Scaling this academic project to all of literature (possibly as its own FRO) would be like Google putting in the up front cost to build the best graph of the web which can then be amortized over cheap and fast queries of that graph as compared to FutureHouse’s approach of conducting searches from scratch with every question. That likely can be done in a cost-effective way given that several academic groups have scaled to millions of papers.
While it’s possible that frontier LLMs that score highly on multiple choice domain knowledge tests may be all that’s needed, it seems likely in the high stakes realm of biological research, a system that actively and painstakingly validates and organizes each fragment of knowledge is important as opposed to a model conducting probability-weighted recall of its weights.
Similarly, with the exponentially rising quantity of biomedical data that largely sits in disparate datasets, strong efforts have been made to integrate the datasets by creating better software infrastructure and automatically annotating experimental context. These pools of data will train foundation models for biomolecules, virtual cells, and tissues to make useful predictions while search engines sitting atop heterogeneous datasets will make natural language queries possible.
Together, progress in these three directions will enable the rapid, systematic relation of novel experimental observations to existing knowledge and data at otherwise unimaginable scales. In the longer term, these resources will be crucial for building highly effective systems for autonomous science, as leveraging prior knowledge significantly boosts their performance, just as it does for human scientists.
Reasoning
A sophisticated autonomous scientist would not only access the continually updating scientific knowledge-base but also be able to reason upon it to come up with novel hypotheses to test, experimental designs to test said hypotheses, what to do next after the experiments end, and how to handle errors along the way.
Science – especially novel discoveries – requires both extreme knowledge compression and advanced decision-making and reasoning capabilities. While the last decade focused on scaling up information compression, we’re now firmly in the era of advanced reasoning models.
I spend significant ink on this section for the reasons below; with that said, feel free to skip if they aren’t relevant for you.
- Reasoning is intuitively and obviously absolutely crucial to human-level or superhuman scientific exploration. So much so that it’s a prerequisite, with most scientific exploration activities “downstream” of reasoning. So understanding the trajectory and time until the cost-adjusted AI performance is sufficient is essential to deciding when to build an autonomous science startup
- After doing all the work for this section, I believe we should expect cost-effective AGI in the next couple years. Therefore, the time to build such an ambitious startup is now.
- While the need for reasoning is intuitive and obvious, the exact mechanism through which AI reasoning occurs is unintuitive and non-obvious. Walking through those details helps understand the drivers of progress
- Those mechanisms (i.e. RL, search, TTT, program synthesis, and multi-agent self-play) are the exact tools that an autonomous science startup will likely need to employ to push the bounds of the SotA
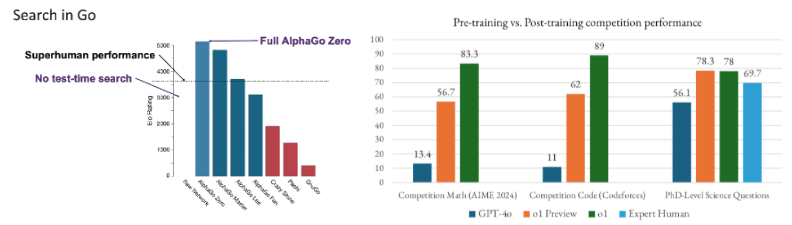
Within the last few months, OpenAI’s o1 and DeepSeek’s R1 replication of it generalized and robustly productized reasoning for the extremely open-ended and unverifiable world of language and chatbots, improving benchmarks almost across the board whereas most prior search algorithms like MCTS work for some domains like Go but not poker. The models can use more compute spend at inference time to “think longer.”
Interestingly, all it took was the simplest possible method of scaling training data and feeding it into a basic autoregressive Transformer. They generate a ton of CoT trajectories in verifiable domains and then use model-supervised deep RL to distill the learnings of how to choose the best path, backtrace, etc. into the base model itself. Only now could such general, open-ended reasoning be built since natural language reward data has exploded with the scaling of RLHF and the base models themselves have gotten pretty good at verifying.
Boy have we learned a lot since o1’s initial release just months ago.
o3 took presumably the same architecture and massively scaled its size. It achieved roughly human-level reasoning on AGI-ARC and solved some of the hardest math problems out there.
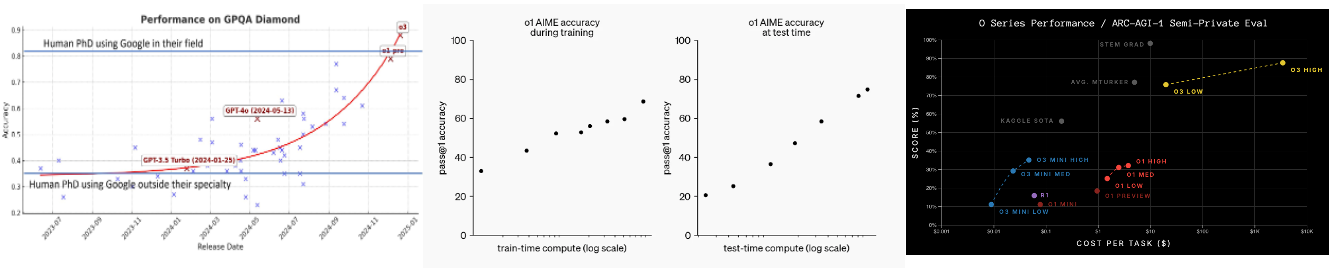
While these models currently require enormous inference cost, they’ll continue to get rapidly cheaper as the CoT datasets become higher quality and the models are trained to find faster trajectories or short-cuts to the correct outcome. In fact, the cost of GPT4 level intelligence fell 1,000x in 1.5 years and the cost of o1 level intelligence fell 27x in the last 3 months (!!).
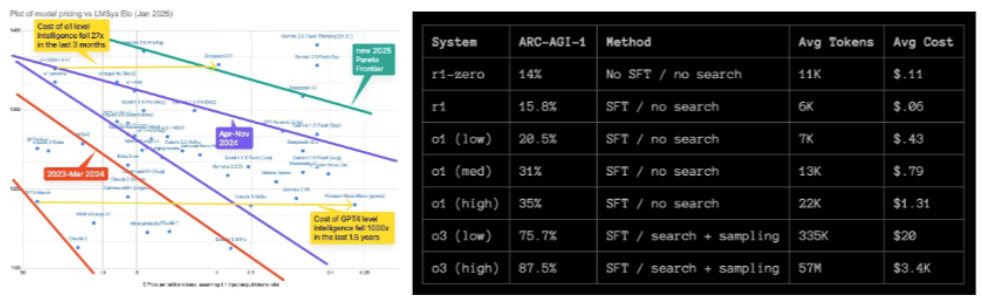
Meanwhile, R1-Zero is the first reasoning model known to have successfully trained from a cold start with no human labeling data to bootstrap à la AlphaZero. Without the human data as guardrails, it exhibits some interesting signs of incoherence and illegibility in its reasoning, however that’s of course what led to move 37.
As Ilya once said, "all examples of true creativity come from RL."
This model to me is far more important than R1 which gets all the media attention, because it serves a PoC pointing to a future with no human bottlenecks, even in the initial data acquisition.
It remains TBD as to how long it’ll be til the models themselves are good enough to serve as verifiers for truly generalized situations and to what extent reasoning traces can be generalized or transfer learned to entirely novel situations especially in domains that are not readily amenable to verifiers. But there's hope on that front as well as RL has been successfully applied to increasingly general scenarios from board games to now Minecraft and robotics.
To get to generalized superhuman reasoning, it may be the case that by scaling CoT training data (which happens naturally every time one of these models is queried), the model can memorize a sufficient number of “reasoning vectors” to pattern match and apply out of distribution.
Alternatively, maybe truly general reasoning will require an architectural advance. One really interesting line of work towards maximal generalization is program synthesis which involves uncovering the program or function that transforms an input to an output by deep learning-guided combinatorial search over discrete operators. It’s the System 1 to LLM’s probabilistic pattern-matching System 2 thinking.
Another exciting method not often discussed that’ll meaningfully impact reasoning in novel situations is test-time training (TTT). Whereas o1’s inference scaling is effectively productized CoT, TTT is effectively productized test-time fine-tuning / LoRA and the authors speculate it could be shipped in frontier models as early as 2026. Models could unify search with learning by having the base model permanently update its parameters in response to the newly learned useful information. This could allow an AI scientist to constantly update itself with useful new information and mental models without having to be retrained from scratch.
Ultimately, some combination of search algorithms, active test-time search, TTT, and program synthesis will be amplified by tool use, multi-agent self-play and collective intelligence, which Noam, Demis, and Ilya all said is the frontier they’re working on.
In addition to reasoning advances, we’re also awaiting the training of step-function larger training clusters in the 100Ks to possibly 1M H100s enabling the jump from GPT4 to 5.
All in all, AI’s scientific reasoning will undoubtedly be profound in the near future, especially considering that o1 Pro already appears capable of being useful or even making novel discoveries in biomedicine and math1,2,3,4.
Hypothesis Generation
When you give a being with strong intelligence and reasoning capabilities access to scientific literature and data, they may just be able to come up with interesting hypotheses to test. While the work in autonomous hypothesis generation is inconclusive with contradictory statements abound, early studies suggest that latest models are already sufficiently capable and certainly with everything described above, will be soon.
One simple problem is how quickly the goal posts move – GPT4o, o1, o1 Pro, and o3 came out in May, Sep, Dec respectively. Given how much these improve in STEM fields and reasoning, any paper before 2024 is largely irrelevant and there hasn’t even been time to study o1 Pro, though an anecdotal demo from an immunology professor suggests a serious inflection point for hypothesis generaiton.
Another issue with current studies is that rigorously evaluating a research idea’s novelty and usefulness is extremely difficult. The only way is controlled and blinded reviews from domain experts at large scales, but that’s very expensive, arduous, and unscalable.
For what it’s worth, the singular study I found that tested a SotA model in such a controlled study with 100+ NLP researchers states that the human experts rated Claude 3.5 Sonnet’s research ideas more novel but less feasible than peer human experts. Other less rigorous studies also suggest their usefulness but some dispute that.
Some other preliminary takeaways include:
- Access to knowledge graphs significantly improves LLM performance
- Utilizing both literature and data improves performance more than either one or the other
- Iteratively updating hypotheses or using multi-agent frameworks can improve quality or diversity
- Even studies that do find that models can generate meaningfully novel hypotheses, the uniqueness of their ideas asymptotes so one couldn’t scale inference indefinitely to continuously generate novel ideas
No one’s created an LLM, no less a LRM, trained from scratch on scientific literature, data, and causal knowledge graphs that’s then guided in post-training towards generating novel, feasible and useful scientific hypotheses. Even if frontier models ingested all that data to achieve their roughly PhD level domain knowledge, a small model trained from scratch without the baggage of the entirety of the internet may prove more effective given speed.
IRL Experiments
The fortunate timing for building autonomous science is that the individual tools needed to measure and edit biology have long been riding exponential cost and throughput curves. One can buy commercially available, off-the-shelf instruments that run the assays at scale and reasonable cost, roughly at the press of a button.
Moreover, all the tools that have historically not been automated like microscopes, cryo-EM/ET machines, MS, etc. are now becoming automated as well thanks to AI. These fields, like compute, will continue to ride their own exponential improvement curves.
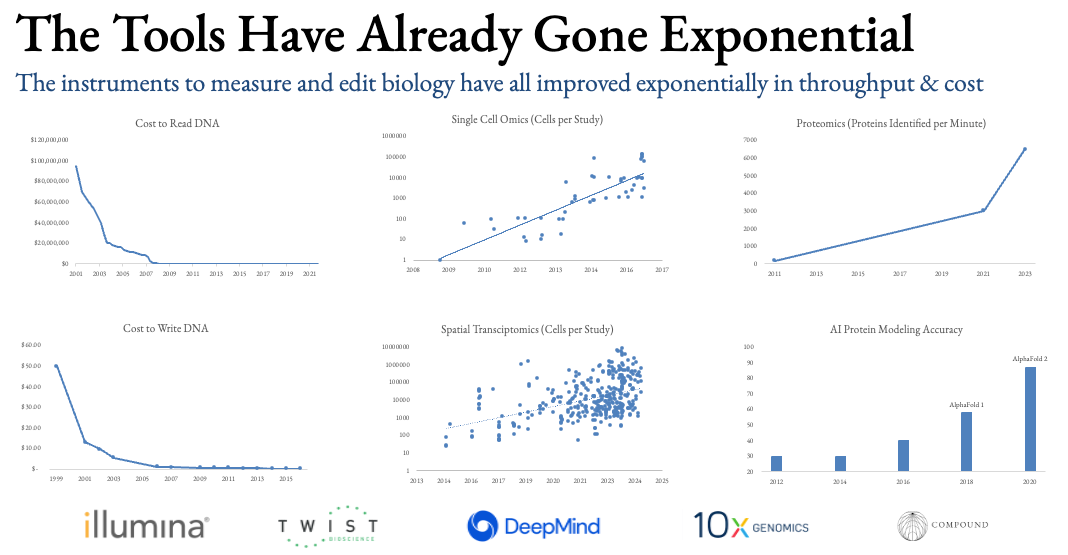
What’s needed now is a robotic system that moves the assays from instrument to instrument to complete experiments in end-to-end automation. For decades, people have toiled away at this, but lab robotics have largely been limited to self-contained boxes for pipetting or unmoving arms. Neither were bestowed with sight so couldn’t tell if a sample was contaminated for example and entirely lacked fluid intelligence so were brittle, tedious to program, couldn’t handle errors, etc.
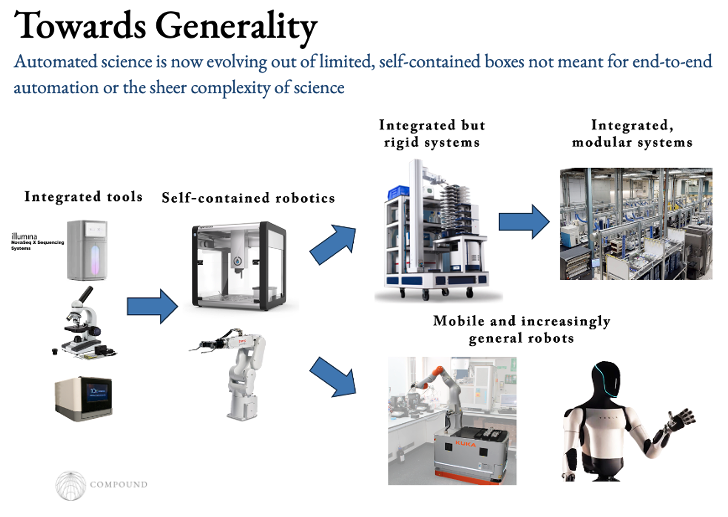
The obvious unlock for orchestrating lab robotics is ChatGPT, which made the once arduous task for the world’s few lab automation specialists as easy as plugging in a natural language interface. Indeed, in a lab automation hackathon I went to, a team made in one day a robotic arm that runs experiments end to end, analyzes its data, decides what to do next, and can be interacted with and programmed in natural language. The latest models are even starting to approach human level performance on trouble shooting and implementing lab protocols.
The physical implementation of experiments has also gotten far easier and more sophisticated. Once it became clear that Transformers were a so far limitlessly general model architecture and scaling them would yield ever more generality, researchers and VCs immediately turned to the next most obvious thing to throw infinite compute and data at: robotics. So we’re getting increasingly general and intelligent robotics with strong visual processing built in. Startups are trying to make smart arms, mobile smart arms, and humanoids suddenly became among the most consensus and crowded areas.
A final, underrated technology that’s arisen in the last few years is Isaac Sim style simulation, which will be vital for making lab robotics maximally seamless. Lab automation specialists spend tons of time manually calibrating robot movements for 3D manipulation. Much of this could be offloaded to a cluster running massively parallel virtual simulations to determine the optimal movement for accomplishing a goal and over time generalize that policy to new environments or tasks.
All in all, these general and mobile robotics systems will be a boon for the low-throughput, discovery-oriented science that demands maximal flexibility. They can be placed into existing labs and run 24/7/365.
Now, to temper expectations, the best automation currently available in our household is a dish washer. It’s reasonable to assume that truly generalized, drop-in, cost-effective robotics won’t be here for a little while, especially given data requirements.
In the meantime, a separate track towards general end-to-end automation that’s been underway for decades and now operating at warehouse scales is progress towards integrated, modular systems. An intermediate step on the journey were companies like HighRes Bio that sell integrated but rigid “workcells” that often place one robotic arm in the middle of 5-10 pieces of equipment. These systems are built to run a fairly narrow set of experiments, can’t be rearranged easily, and can’t run multiple experiments in parallel.
Today, Emerald Cloud Lab, Gingko, and others have 100Ks sq ft in warehouse space with a web portal for scientists to run just about any experiment they want from their laptop. Remote researchers or autonomous agents can select from a list of protocols and run the desired experiment at the click of a button. It also enables the dynamic scaling up of equipment use as needed and experiments to run in parallel. The key for these high capex businesses, like AWS or TSMC, is maximal utilization and uptime.

Pharma is starting to catch on to this approach and are hiring these cloud lab orchestrators to help them centralize all their equipment from their many disparate labs into a few massive warehouses, ensuring far less equipment redundancy and better utilization.
The bull case for these large-scale cloud labs is that, like other capex heavy businesses, if they can keep expanding their top-line and userbase, a self-reinforcing dynamic may evolve. Continually reinvesting that rising revenue back into their infrastructure yields compounding improvements in cost cutting and offering quality – in diversity of experiments handled, speed, and ease of programming. This drives yet more demand and the ability to amortize those costs over an ever-greater userbase.
Maybe sufficient industry adoption may even encourage the equipment makers like Hamilton to redesign their instruments from the ground up for this automated pipeline instead of being built to serve humans. Out with 96 well-plates and human legible processes. Maybe instruments built for that future are an opportunity for startups.
We estimate the current total market spend on cloud lab services and infrastructure build out to be very roughly on the order of $200M.
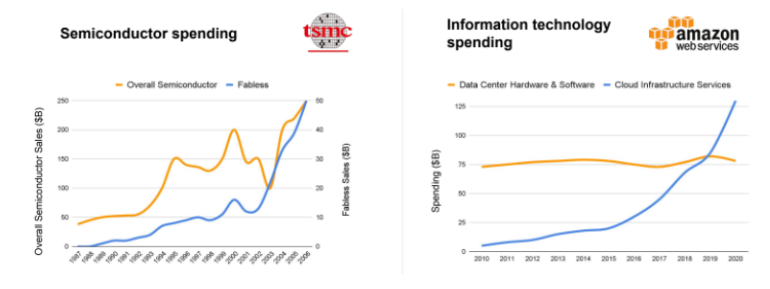
Maybe the demand for centralized cloud labs doesn’t materialize and an opposing future emerges in which smaller scale purpose-built CROs pop up all over the place that specialize in solving relatively narrow tasks. These startups identify experiments like enzyme engineering or whole-plasmid sequencing that 1) industry needs to run in large volumes, 2) have a high Idiot Index, and 3) are too complex or take up too much physical space to automate via desktop instruments mentioned at the beginning of this section. They then design a way to do it from the ground up at higher throughput and lower cost, often with the help of chips or miniaturization via microfluidics.
Alongside these task-specific CROs in this third vision of the autonomous future, the extremely long tail of lesser funded academic labs who can’t afford fancy autonomous equipment build maximally cheap and composable hardware from open source blueprints according to the spec below.
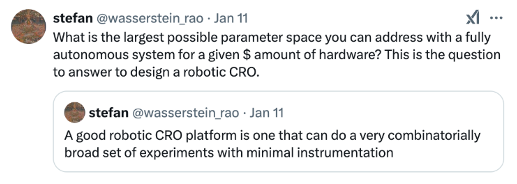
In either the centralized or decentralized lab automation scenario, every experiment run through automated pipelines adds to the code base for orchestrating the robots. Each experiment is the combination of sub-tasks like GoldenGate cloning, yeast transformation, and plate reading. So as libraries of sub-tasks and experimental designs proliferate, running a new experiment will become as easy as no-code, drag-and-drop configurable building blocks in the next several years. After that, maybe AI systems will become capable enough to design protocols themselves.
In Silico Experimentation
There’s a computational tool to predict and measure every step in drug development. The field’s particularly crowded on the preclinical side with many approaches for structural modeling, small molecule, antibody, macrocyclic peptides, toxicity and off-target effects prediction, cellular response.
These models can currently be thought of as in silico high throughput experiments to narrow down the search space as much as possible to then verify the promising leads in the lab.
AlphaFold is the first to converge on experimental accuracy, meaning its predictions are nearly as accurate as running the full test in the lab. The 40-year overnight success is of course ahead of less mature areas like NNPs, virtual cells, and the various de novo generative design tools, but they’ll get there in 5-10 years.
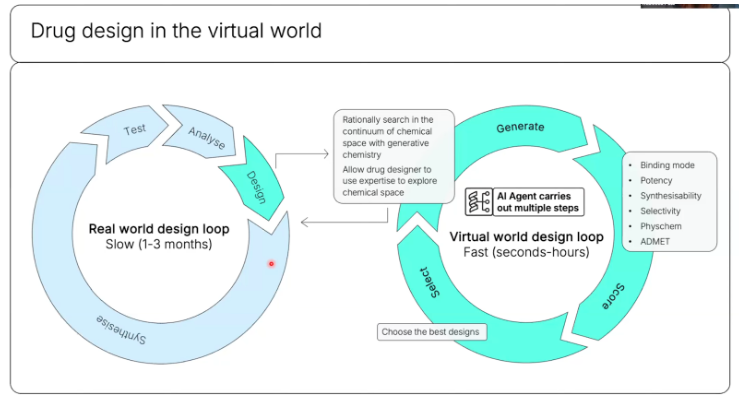
As both search and experiments can increasingly be conducted entirely within a computer where the feedback loops are tight and can be arbitrarily parallelized, it will become possible to train a superhuman model on the meta of doing science itself.
The physical world will forever be too slow and unscalable for that purpose. For context, AlphaZero played itself 44 million times over in its training. And chess is infinitely simpler than scientific discovery.
So while continued exponential improvement in measurement instruments’ throughput combined with increasingly general purpose robotics will help expedite and automate specific protocols, it likely won’t be enough to train a model on how to do science in general.
To get a glimpse into how this brave new world may look and how different the biological sciences may look, consider the perspectives below from a virtual cell startup and Isomorphic, respectively. First, advanced mechanistic interpretability techniques are used to digitally dissect the virtual cell to find a drug target to launch a drug discovery campaign.
Suppose we have a bunch of single-cell perturbation experiments and are trying to solve the problem of finding the perturbation a* that moves your cell from starting expression state s0 to a desired end expression state s*. How might your virtual cell combined with mechanistic interpretability techniques speed this up?
You noise the data sufficiently, run a forward pass, and look at the major SAE features that are active across tokens in the cell. This gives you a general idea of where you’re at in cellular state space and what the major biological modules at play are.
Then, you might start doing some cheap attribution methods like the forward-gradient method or DeepLIFT, which require O(1) forward and backward passes, to get an idea of which input genes matter for the output.
Then you might try a more expensive sparse feature attribution method, either computing attribution of output logits (particular genes you’re interested in) or the activation of other features with respect to upstream features. The cheapest way of doing this requires 2 forward passes for each of the features you want to run attribution on, but could run more forward passes to get a more precise estimate. With such a feature-feature or feature-logit attribution graph, you can begin to get an idea of which features are most causally central to the cell’s computation.
Then you might try to see which of these features is most important in mediating the cell state transition by ablating them from the model one-by-one. Supposing you identified a small group of candidate mediator features, you might then further ablate features downstream of them to determine the feature pathways through which they exert their effect on the outcome state, a kind of extended description of the perturbation’s mechanism of action.
Eventually, using a combination of attribution, ablation, patching, and other methods, you might reverse-engineer circuits inside your virtual cell for the minimally sufficient perturbation needed to cause some desired change while minimizing undesirable effects – like activation of the “toxicity feature”.
This is what true, novel, entirely in silico target discovery may look like.
Now, let’s say it successfully identifies a disease-causing protein. Even the current capabilities of frontier labs like Isomorphic can generate promising drug leads in the span on weeks. A talk by one of their senior scientists gives an unusually clear look into their process and one can imagine how it’d fit contiguously with the future virtual cell work.
They’d predict the atom-level 3D structure of the protein with experimental accuracy using AF3, replacing the years-long cryo-EM crystallization process with a prediction process currently measured in a few minutes.
The generation of chemical matter to drug that structure would be guided by a range of parameters like binding affinity, diversity of matter, and when connected to the virtual cell, it could also be guided by correlations with feature activations like toxicity.
Isomorphic then ranks all these compounds and can currently generate such predictions in under five seconds so they can score millions of compounds over the course of a few days. They take the top scoring compound into the next phase of virtual design and generate alternative versions with edits informed by the prior round.
They continue this loop until they’re unable to make further improvements to the scores.
The advent of NNPs enables teams to then take the final compounds, place them into 3D atomistic simulations of the real intracellular environment, and “press play.” Watching how the compound interacts with the protein and its immediate local surroundings over time may identify time-dependent events affecting drug efficacy like emergence of cryptic pockets or metastable conformational changes.
The team could then take the remaining optimized leads and run them through the virtual cell, using it to predict toxicity or off-target effects as well as the mechanistic interpretability tools spelled out to glean additional information.
Once no more optimizations can be made in silico, the drug hunters finally enter the real world, calling the cloud lab API to synthesize the top hits and run the corresponding tests using our ever improving options for models of human biology.
I went into this detail not only to provide a glimpse of how we at Compound believe drug discovery may look in just ten years but because it’s crucial to remember just how many discrete steps and decisions are required. Each step is a branching point in which an AI model conducting autonomous science would need to decide what to do next amongst the infinite options.
With real world experiments measured in days to months, this entirely in silico approach is the only feasible way to move towards teaching an AI to navigate the infinite labyrinth of decision trees and learn “the meta of doing science.” Just like Isaac Sim is utterly essential for general robotics, computer-based exploration is essential for learning to do science.
Ultimately the goal is to build an end-to-end system where you have granular step-level data, outcomes, and can run experiments massively in parallel. You then do deep reinforcement learning to update the decision policy after each experiment to make better decisions next time.
I can imagine a future (that isn’t decades away) in which the entire combinatorial path described above from basic science and target discovery to lead optimization happens in seconds to minutes and where biology labs are running such experiments in the millions at a time in parallel in multi-gigawatt datacenters.
That is how you learn the meta of doing science itself.
Proofs of Concept
Several dozen PoCs from academic labs, startups, Big Pharma, and Big Tech have demonstrated that it’s indeed already possible to stitch together the five components laid out above to conduct end-to-end autonomous science in both biology and materials science across a long list of subtopics from systems biology to photovoltaics to 3D printing to antibody development.
Some exciting and emblematic examples are below:
- The first novel knowledge discovered by a closed-loop autonomous system in biology came from Adam in 2010 which inferred gene functions in S. cerevisiae through gene deletions and auxotrophic analyses
- To date, the most impressive demonstration of a self-driving lab is A-Lab, which is a collaboration between Lawrence Berkeley National Laboratory and DeepMind that has carried out 3,500+ chemical synthesis experiments fully autonomously over the last year.
- A systems biology experiment that hopes to demonstrate for the first time that an area of science can be investigated using robot scientists unambiguously faster, and at lower cost, than with human scientists. It will initiate and execute in parallel one thousand hypothesis-led closed loop cycles of experiment per-day on μ-bioreactors integrated with mass spectrometry.
- A creative framework for multi-agent collaboration where the agents were given different roles and expertise in a particular discipline — immunology, computational biology, machine learning. A PI agent instructed them to design novel nanobodies to target SARS-CoV-2 and, in partnership with a human researcher, had conversations with them about approaches to the problem and assigned them tasks to complete. Using tools like AlphaFold, they designed 92 nanobodies, 90%+ of which bound to the original variant and two bound to new variants.
Conclusion
Only after spending many hours researching, talking, and writing am I convinced that it’s actually possible to begin tackling as ambitious a goal as automating science.
In fact, we at Compound will take the under on the Nobel Turing Grand Challenge, which sets 2050 as the goal for making a Nobel-worthy discovery entirely autonomously.
Now that we’ve detailed what tools you can build with and our argument for why now is the time to build, we next will talk through what types of business models can be employed and what commercialization approaches we’re most excited about in Part II.