Organ-on-a-Chip: Simulating Human Biology at Scale (The Century of Biology: Part I.V)
The rate of progress towards curing a given disease is a function of how quickly scientists can run experiments. Currently, it takes 10+ years to reach a definitive answer on whether or not a drug addresses an issue in human beings. Compare that to a tech startup, which is constantly iterating through different ideas, throwing them against the wall and seeing what sticks. A reason that industry has moved so quickly is computer science lends naturally to failing fast.
Curing diseases in mice is closer to the tech industry’s ability to experiment and rapidly iterate, because researchers can just try things at will. However, ultimately, there’s no true substitute for testing in humans. The lacking effectiveness of our current models of human biology explains why 90% of drugs fail during human clinical trials after passing animal and other models. Pharma companies narrow down a massive array of possibly useful drug targets to clinically test only one. Each failure costs millions of dollars and years of clinical work, making the selection a terribly risky bet. Approximately 30% of those failures are due to toxicity, which can have tragic consequences.
New technologies like organ chips are aiming to replicate human biology as faithfully as possible, allowing scientists to run effective simulations of real-world clinical trials at scale.
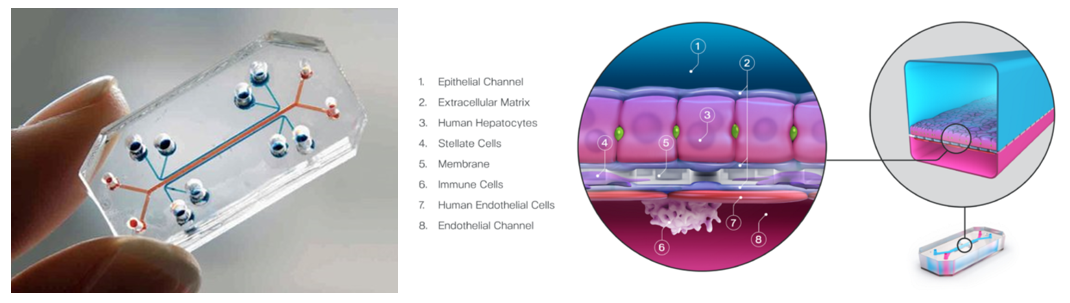
Organ chips are millimeter-sized replications of specific organs or tissues using human cells (iPSCs, patient-derived or standard) to more faithfully or safely model human physiology and pathology for both disease modeling and basic science. All of these devices feature hollow channels surrounded by living cells and tissues that are cultivated under continuous flow of fluids. Some mimic organ-level structures like tissue-to-tissue interfaces, while others offer essential biological mechanics like breathing or involuntary muscle contractions. For example, the image above on the right visualizes a liver chip that includes four different cell types, vascular blood flow, and oxygen flow to model drug toxicity and effectiveness.
The cell channels are generally housed in clear plastic and thus allow for real-time, high-resolution microscopy. It's also generally possible to incorporate a range of sensors integrated within the device to observe tissue health and function, including real-time oxygen level tracking47, alterations in tissue barriers, 49,50 and cellular electrical activity. 51
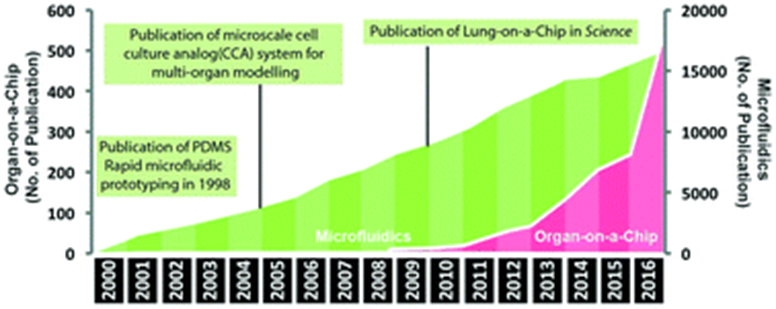
The development of organ chips began in the 90s following the progress of stem cells since the 60s and microfluidics since the 80s. In 2010, the field reached a major milestone: researchers at Wyss created the first microfluidic device to capture complex organ-level physiological and pathological responses, not just maintain cell- or tissue-level differentiated functions. They built a highly sophisticated lung model:
One channel was lined with a layer of primary human cells isolated from the lining of the lung air sac, or alveolar tissue, which after a couple of weeks multiply and differentiate into the cells native to alveolar tissue. These secrete mucus when cultured on the top of the permeable membrane and exposed to air introduced into the upper channel. Human capillary blood vessel cells from the lung were used to line the lower surface of the same membrane with a liquid medium flowing through the lower channel to provide nutrients to both the alveolar and capillary cells as they would in the body. Two hollow chambers were placed on either side of the two microchannels which allowed for cyclic suction to be applied, thereby enabling the rhythmical stretch and relaxation of the engineered 'alveolar-capillary interface' to model the mechanics of breathing. The device provides a means to simulate the effects of air pollution and bacterial infections on the lungs, and to investigate what happens when lungs become inflamed (Huh, Matthews, Mammoto). The complex breathing action of the engineered lung also subsequently opened up new insights into the cause of pulmonary oedema, a serious condition caused by excess fluid in the lungs, and helped predict the activity and toxicity of a new drug developed by GlaxoSmithKline for the disease (Huh, Leslie, Matthews; Kusek).
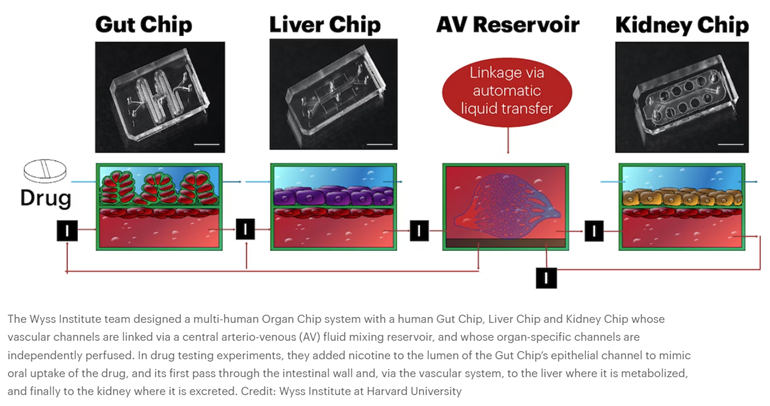
Then, in 2012, DAPRA requested grant applications for a multi-organ system that combines 10 different organ chips to create a functional human body-on-a-chip, fluidically linked together. Wyss successfully completed the task, combining their system with computational modeling and experimental data generated from the platform to predict human drug PK/PD. “We serially linked the vascular channels of eight different Organ Chips, including intestine, liver, kidney, heart, lung, skin, blood–brain barrier and brain, using a highly optimized common blood substitute, while independently perfusing the individual channels lined by organ-specific cells. The instrument maintained the viability of all tissues and their organ-specific functions for over three weeks and, importantly, it allowed us to quantitatively predict the tissue-specific distribution of a chemical across the entire system,” said an author on the resulting Nature studies.1,2
At this point, practically every organ has been simulated, and the devices have been used to model diseases ranging from Parkinsons to cystic fibrosis to COVID. Many have achieved the gold standard for something aiming to simulate human organs: replicating the findings of in-vivo clinical studies. Some results have been included in applications to the FDA to move new drugs into the clinic.
The spreadsheet below catalogues many of the individual organs simulated as well as the body-on-a-chip multi organ systems.
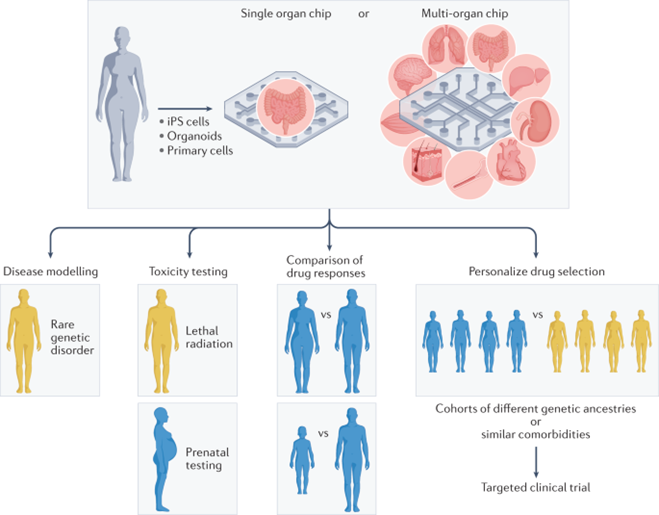
As the technology has improved, its use cases and effectiveness at each have become clear. The section below details each use case and provides an example of a state of the art organ chip being applied in that way:
Simulating biology as is to study physiology and disease pathways:
- One of the most clinically relevant uses so far for lung chips have ben to study respiratory virus infections, particularly COVID:
When these chips were infected with a pseudotyped SARS-CoV-2 virus and perfused with clinically relevant doses of the antimalarial drugs chloroquine or hydroxychloroquine, these drugs were found not to be active, thus predicting the negative results seen in clinical studies. By contrast, a related drug, amodiaquine, was shown to actively inhibit SARS-CoV-2 pseudovirus virus entry on-chip, as well as infection by native infectious SARS-CoV-2 in a hamster model. Importantly, these results contributed to the repurposing of this drug and its entry into clinical trials for COVID-19 in Africa, where this drug is used widely for antimalarial prophylaxis.
- Liver chip with exposed to 200 living bacterial species with similar complexity as the gut microbiome replicated the mechanisms of inflammatory bowel disease as observed in the clinic.1
- Sometimes new fundamental discoveries are made:
Revelation that physical forces associated with cyclic breathing motions influence lung innate immune responses to viral infection59. This work led to the discovery that receptor for advanced glycation end products (RAGE), an inflammatory mediator that is most highly expressed in the lung alveolus in vivo, has a central role in this response. Moreover, when a RAGE inhibitor drug (azeliragon) was tested, it potently suppressed production of inflammatory cytokines and synergized with the antiviral drug molnupiravir in this model. These results were included in a recent submission of a pre-IND (Investigational New Drug) meeting application to the FDA to request initiation of a Phase 2 clinical trial to test whether azeliragon can suppress the cytokine storm in patients with COVID-19.\
Simulating possible future scenarios to devise new therapies:
- The ability to model virus evolution using lung airway chips61 offers the possibility of predicting new strains before they emerge, which could accelerate development of both drugs and seasonal vaccines for various viruses (for example, influenza, SARS-CoV-2 variants) in the future.
- Given the ability of organ chips to reproduce human bacterial infections in tissues from both healthy individuals and patients with chronic diseases (for example, cystic fibrosis)11,40,43,73,74, and to mimic fungal infections91 as well, similar approaches may be used for the discovery of antibiotics for emerging antibiotic-resistant organisms.
Studying phenomena that cannot be replicated by other models:
- Some diseases can’t be effectively modeled in animals like liver toxicity and Alzheimer’s
- Vetting biologics designed specifically for humans: 40% of new drugs are biologics, many of which cannot be tested on animals. Biologics can trigger severe immune reactions [Peterson et al., 2020; Kloks et al., 2015].
- Studying potentially dangerous therapies: E.g. potentially lethal radiation exposures
- One study modelled the cancer tumor microenvironment, which can’t be easily modeled in conventional cultures
- Studying rare genetic diseases:
A kidney chip replicated phenotypic features of two rare genetic disease: Knocking down OCRL expression in the cells revealed that proximal tubule cells lacking OCRL upregulate collagen deposition, which can contribute to the interstitial fibrosis and disease progression seen in these disorders. The ability to build chips with patient-derived cells also opens up new approaches for therapeutic development, particularly for patients with rare genetic disorders.
Screening for a drug’s toxicity:
- A blood vessel chip lined by vascular endothelium and perfused with human whole blood reproduced the clinical thrombotic toxicity of a therapeutic monoclonal antibody drug (Hu5c8) that had been previously withdrawn from clinical trials owing to unexpected life-threatening complications that were not detected during preclinical animal testing3.
- A bone marrow chip replicated observed human clinical toxicity in response to cancer therapeutics as well as radiation treatments:
This chip reconstituted differentiation and maturation of multiple blood cell lineages over 1 month in culture and replicated myeloerythroid toxicities in response to clinically relevant exposures to the cancer therapeutic 5-fluorouracil as well as to γ-radiation. Even more impressively, it was able to replicate regimen-specific toxicities of a cancer drug observed in human clinical trials when the drug exposure (pharmacokinetics) profiles previously measured in patients were recreated on-chip. And again, these responses were not observed when the same haematopoietic cells were placed in static cultures.
In addition, personalized bone marrow chip models of a rare genetic disorder, Shwachman–Diamond syndrome (SDS), were created in this study using CD34+ cells obtained from patients with SDS. These chips replicated key haematopoietic defects seen in patients and led to the discovery of a previously unknown neutrophil maturation abnormality in a subpopulation of these patients.
- An intestine chip model of acute radiation injury also replicated the same radiation dose sensitivity observed in humans, which differs from that observed in animal models75.
- Lung chip used to asses safety of T-cell cancer therapy, effectively reproducing non-human primate toxicities of targets under clinical development (link): “Importantly, when a lower-affinity T cell bispecific antibody that was found to exhibit lower toxicity in the lung chip was tested in NHPs, none of the animals experienced the lung inflammation observed with the original higher-affinity antibody, thus confirming the safer profile predicted by this human organ chip. The ability to visualize these responses at high resolution yielded insights into a novel toxicity mechanism in which one drug was found to unexpectedly target the endothelium, rather than a parenchymal cell.” (link)
And drug effectiveness and optimal dosing:
- Fluidically linked body-on-a-chip system can provide testbeds for scientists to conduct complex tests for drug disposition, ADME and PK/PD behaviors. Moreover, the ability to quantitatively predict human drug PK/PD parameters in vitro could be used to optimize dose regimens for new therapeutics and thereby shorten phase I clinical trials.
Microfluidic systems composed of multiple culture chambers in which medium is flowed directly from one parenchymal cell compartment to another via a single channel have been used to carry out PK/PD modelling in vitro for many years, although established cell lines were usually used in early studies33. Later versions separated the flow chamber from the culture chambers with an intervening porous membrane or used transwell inserts as culture chambers26,27,28,29,30 (Fig. 2a). A body-on-chip system of this type composed of linked liver, kidney, intestine, skeletal muscle and BBB chips generated predictions of drug metabolism and barrier penetrance that were consistent with clinical data112. But it is only recently that this type of modelling has been carried out in organ chips lined by primary human cells that are fluidically linked by flowing or transferring a common blood substitute medium through endothelium-lined vascular channels that are separated from parenchymal channels or chambers, as occurs in our bodies13,31,32,34,35 (Fig. 2b,c). This more physiologically relevant approach has been used in combination with computational modelling to generate predictions of drug PK/PD parameters (Fig. 3) and toxicities in many studies (Table 2). For example, it has been possible to quantitatively predict human clinical pharmacokinetics parameters using a multi-organ chip first-pass model composed of linked two-channel human liver, kidney and intestine chips along with an AV reservoir in combination with computational physiologically based pharmacokinetics modelling35 (Fig. 3b). Successful in vitro to in vivo translation of pharmacokinetics parameters closely mimicked results observed clinically (Fig. 3c) and this was carried out using two drugs (cisplatin and nicotine) administered via two different routes (intravenous versus oral). In addition, when the intestine chip was replaced by a bone marrow chip, predictions of cisplatin pharmacodynamics also matched previously reported patient data35.
Selecting more relevant groups for clinical trials:
- Organ chips could also prove useful for validating potential drug targets identified by genomic and proteomics so they can enhance the drug discovery process. This is particularly important because of the great variability found between different ethnic and genetic groups in their responses to therapy (Ingber 2022). Organ chips lined by patient-derived cells would be particularly useful when toxicities are observed in a particular patient subpopulation in phase I trials (for example, women versus men, or a genetically similar subgroup).
Personalized medicines based using patient-derived cells:
- For example, small-molecule inhibitors of cyclin-dependent kinases 4 and 6 (CDK4/6) were identified as potential novel immune checkpoint inhibitors using a chip in which patient-derived tumour cell spheroids containing autologous infiltrating immune cells were cultured within 3D ECM gels surrounded by microfluidic channels99. These CDK4/6 inhibitors augmented the response to PD-1 blockade, thus providing a rationale for combining these agents with other immunotherapies to enhance therapeutic efficacy.
Current industry snapshot:
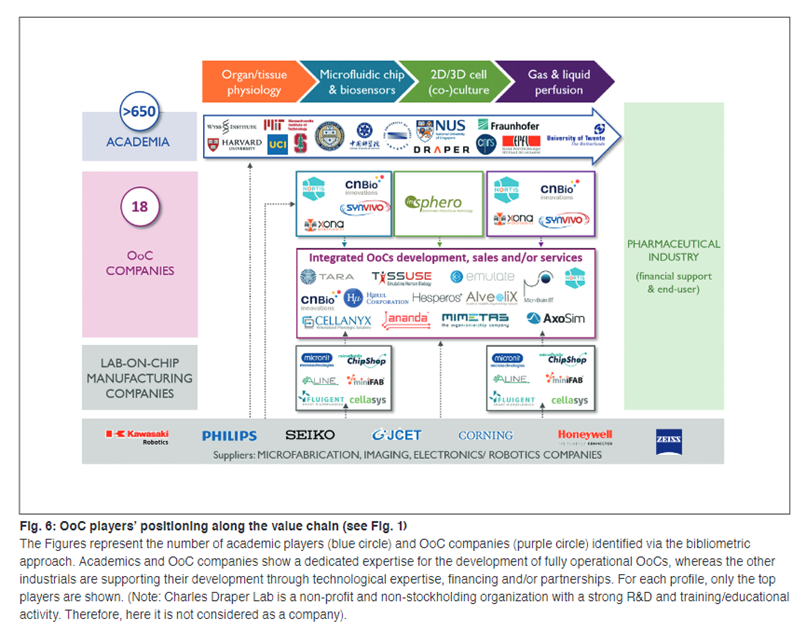
A value chain has developed around this burgeoning industry, with at least $500M raised by startups from VCs to date. Below is Emulate’s body-on-a-chip machine that integrates the various chips automatically. Users can adjust the microenvironment the chips are exposed to via the touch screen.

Estimated impact on drug discovery:
A 2019 survey of 15 pharmaceutical companies forecasted that organ chips could cut R&D costs by 10–26%. Echoing this, a 2022 Nature paper demonstrated that the Emulate human Liver-Chip was far more predictive of drug-induced liver toxicity than animal and spheroid models. The team estimated that the chip could yield more than $3B in excess productivity for the drug development industry, thanks to the model’s improved predictive validity.
What’s next?
All the models listed above are fantastic proofs of concept for each targeted disease model, but to be accepted in large scales by drug developers each chip must be rigorously tested by replicating the same exact experiment hundreds of times until the method’s superiority over existing workflows is statistically undeniable. This exercise has already been successfully carried out for the Emulate Liver Chips. A study involving nearly 800 chips made from two cell donors met the standards set by an independent regulatory body. The chips’ correctly identified nearly 9 out of 10 toxic drugs whereas hepatic steroids exhibited 47% sensitivity and animal models 0%. Moreover, it didn’t falsely identify any drugs as toxic. As a further indication of Emulate’s progress towards integrating their chips into pharma workflows, more than 150 labs including 17 of the top 25 global pharma companies have installed their chips.
Second, there’s been meaningful regulatory progress in the last couple years. The groundbreaking FDA Modernization Act 2.0, signed into law December 2022, explicitly legalized the use of organ chip models as a part of new drug applications. This ruling nixed the FDCA mandate, in place since 1938, requiring all new drugs be tested on animals before human clinical trials. Additionally, the FDA & big pharma collaborated to create criteria to evaluate and adopt organoids and microchips, detailed in this paper. Finally, the FDA’s Innovative Science and Technology Approaches for New Drug division, which has established similar qualification criteria for drug development tools including organ chips, now also offers a defined path to expedite new technology’s acceptance for regulatory use, making it easier for widespread approval once the technology is proven commercially.
Ultimately, commercial adoption will likely occur in stages. Situations like the Liver Chip study described above where current models are known to be sorely inadequate, and the organ chips can effectively replicate human pathology. As the devices’ sophistication, cost, consistency, and overall predictive power continues to improve, biopharmaceutical companies will integrate them into more and more of their workflows, regularly applying them to the full suite of use cases cataloged above.
Read next section: artificial intelligence